Researchers explore how AI-driven tumor-infiltrating lymphocyte (TIL) assessments outperform manual methods in precision but stress the need for robust datasets to ensure reliability in clinical practice.
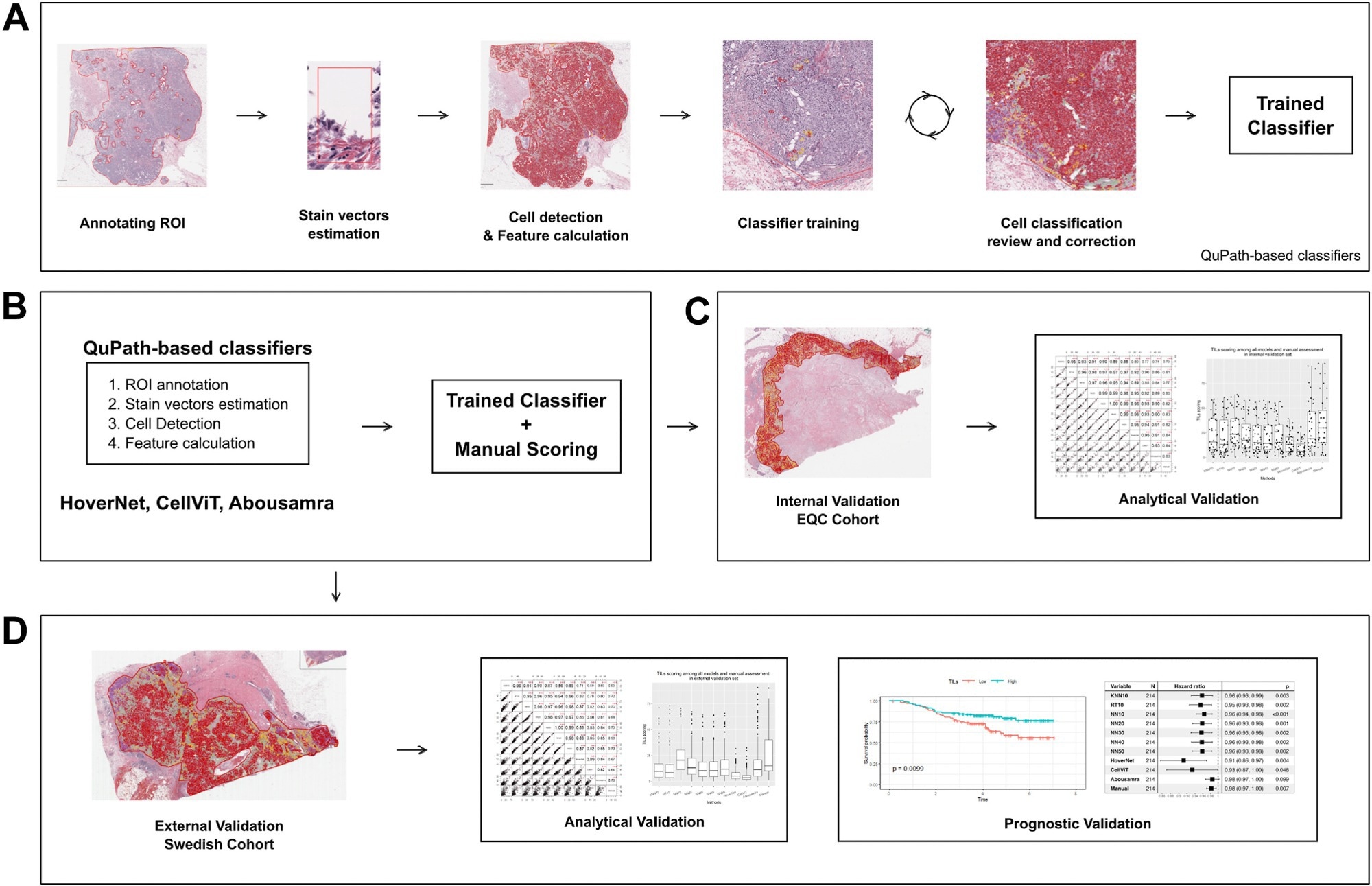
Digital image analysis flowchart for classifiers development and utilization. (a) Preprocessing and classifiers training pipeline (KNN10, RT10, NN10, NN20, NN30, NN40 and NN50). (b) Application of TILs models. (c) Analytical evaluation of the classifiers on the Yale internal validation set. (d) Prognostic evaluation in an independent validation set. Note that the “trained classifier” applied in sub-figures b-d is the one created in a, in addition to HoverNet, CellViT and Abousamra’s. Study: The analytical and clinical validity of AI algorithms to score TILs in TNBC: can we use different machine learning models interchangeably?
In a recent study published in the journal eClinicalMedicine, researchers compared the prognostic and analytical performance of artificial intelligence (AI)-based tumor-infiltrating lymphocyte (TIL) assessment models in triple-negative breast cancer (TNBC).
The past few years have witnessed an unprecedented development of novel treatments for early-stage breast cancer. However, a systematic implementation of biomarker-based risk stratification is urgently needed to prevent under- or over-treatment and select patients who may benefit from additional treatment.
Further, the quantity of lymphocytes infiltrating the tumoral stroma, viz., stromal TILs (sTILs), is a prognostic feature of early-stage TNBC. While guidelines have been released to standardize sTIL assessment, interobserver variability is inevitable, and TIL scoring is limited in its capacity to capture the complexities of the tumor microenvironment (TME).
This underscores the need for advanced, automated approaches that can address variability and provide deeper insights into the tumor-immune interactions. Moreover, the performance of AI models across diverse imaging platforms and datasets remains a critical consideration for clinical adoption.
About the study
In the present study, researchers compared the prognostic and analytical ability of 10 AI-based TIL assessment models. Whole-tissue section slides (WTS) were obtained from 106 females with primary invasive TNBC tumors between 2012 and 2016. Ninety-two slides of 79 patients were used for training and internal testing of models. Further, WTS and clinical data of 215 TNBC patients (from another cohort) were used for external validation.
Automated TIL scoring algorithms were built using the QuPath platform. Models from three families, neural network (NN), K-nearest neighbor (KNN), and random trees (RT), were trained on a subset of 10 images. Additional training scenarios included increasing the number of patient samples (20, 30, and so on). Each method was represented as “MN,” where M was the method’s name (e.g., KNN) and N was the number of training samples.
Each image comprised manual annotations for about 450 cells, with at least 150 lymphocytes and 150 tumor cells. The remaining cells were stroma or other subtypes. Researchers employed a “human-in-the-loop” strategy for model training, involving iterative manual annotations and accuracy checks to achieve optimal classifier performance. This method ensured that the models accurately reflected the heterogeneity of TILs.
In addition to these models, three advanced deep-learning methods—CellViT, HoverNet, and Abousamra’s model—were included to provide a comparative analysis of state-of-the-art techniques. Digital TIL scores were calculated using the easTILs formula for all models except Abousamra’s.
For Abousamra’s model, the percentage of invasive cancer regions predicted as lymphocyte patches was used as TIL scoring. The correlation between pathologists’ manual sTIL scores and digital sTIL scores was determined using Spearman’s correlation coefficient.
Univariate and multivariate Cox regression assessed the prognostic value of TIL scores adjusted for age, histological grade, nodal status, and tumor size. HoverNet and CellViT models were pretrained on the PanNuke dataset, which encompasses over 200,000 nuclei across 19 tissue types, enabling these models to achieve fine-grained cell segmentation and classification.
Findings
The team developed seven models (KNN10, NN10, RT10, NN20, NN30, NN40, and NN50). In the internal validation set, RT10 and KNN10 had the widest distribution of TIL scores, while NN models had comparable and consistent distributions. By contrast, CellViT and HoverNet had the narrowest distributions, while manual scoring and Abousamra’s model had the widest distributions. The correlation of digital sTIL scores with manual sTIL scores varied across models.
RT10 showed the best correlation among models trained on limited samples; KNN10 exhibited moderate correlation, and NN10 showed a slightly better correlation. Increasing the number of samples gradually increased correlations. CellViT and HoverNet showed the second-best correlation. However, significant disparities emerged between internal and external validation sets, with all methods showing reduced performance in the external cohort.
Differences in imaging platforms—Yale’s Leica Aperio system versus SCAN-B’s NanoZoomer platform—likely contributed to these discrepancies. In the external validation cohort, TIL score distributions were much narrower for all methods; all correlation coefficients decreased in value.
Notwithstanding, RT10 still exhibited the best correlation, while KNN10 had the lowest correlation. Moreover, increasing the sample size did not improve the correlation, unlike in the internal cohort. Further, associations of models and patient outcomes were investigated in the external validation cohort, with invasive disease-free survival (IDFS) as the clinical endpoint.
IDFS was defined as the time from diagnosis to death from any cause or breast cancer-related events. In univariate Cox regression analysis, all except Abousamra’s model showed significant results and had similar and overlapping hazard ratios.
The multivariate analysis yielded similar results for all models, albeit CellViT and Abousamra’s model had borderline non-significant results. The study noted that continuous TIL scoring provided more robust prognostic analysis than cutoff-based scores, given the variability in distributions across methods.
Conclusions
In sum, the researchers evaluated the prognostic and analytical ability of 10 AI-based TIL models against IDFS. Seven models were developed and three were pretrained, validated models.
Regarding analytical performance, AI models achieved moderate to good correlation, even when trained on more samples, albeit models of similar architecture (e.g., NN10-50) had a high correlation. Nonetheless, the study highlights the persistent gap between internal and external performance, emphasizing the need for rigorous external validations.
Their performance dropped in the external cohort; increasing the training sample size did not improve the correlation. Nevertheless, the prognostic potential of digital TILs was notable for nearly all models, even for models with smaller training sample sizes.
The study also stressed that for clinical adoption, AI models must offer transparency and explainability, allowing clinicians to understand and trust the predictions. This includes the ability to review misclassified cells directly in the segmentation output.
Overall, the researchers stress the importance of large, diverse, multi-center datasets to serve as benchmarks for standardizing and validating AI models. These datasets are essential to ensure clinical compatibility and eliminate the risks associated with model-specific biases.
Source link : News-Medica